NRPS tailoring domains
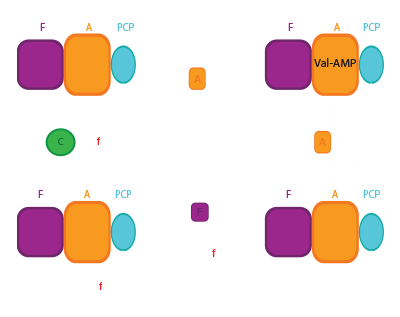
Relevant Schmeing lab papers: Fortinez et al, Nature Comm 2022; Fortinez at al, JACS 2022; Alonzo et al, Nature Chem Biol 2020; Reimer et al, Science 2019; Reimer et al, ACS Chem Bio 2018; Reimer et al, Nature, 2016; Alonzo et al, PLOS One, 2015