Research

Structure and Function of Macromolecular Machines
The general goal of the lab is to understand how some of the large enzymes in the cell act to perform their important functions. These enzymes often require supramolecular organization and complex architecture to function. For example, both the ribosome and some nonribosomal peptide synthetases use more than 100,000 atoms to make peptide bonds, while the proteases that break these bonds can be very small. Of course, these assemblies require regulation, processivity and fidelity, which contribute to their increased size. Our lab investigates both the manner by which cellular machines achieve these roles, and the mechanisms of their principal functions. To do this, we combine X-ray crystallography, electron microscopy and biochemical techniques.
Nonribosomal Peptide Synthetases
Nonribosomal peptide synthetases (NRPS) are large macromolecular machines that, like the ribosome, catalyze peptide bond formation. Instead of making proteins, these enzymes produce a large variety of small molecules with important and diverse biological activity.

NRPS amide bond forming domains
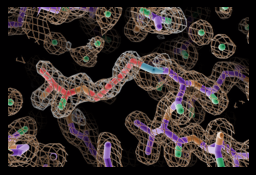
In basic NRPS modules, the C domain catalyzes the key catalytic event of NRPS function, amide bond formation to link substrate monomers and grow the peptide chain. The C domain is a pseudodimer of ~450 amino acids, with N- and C-terminal subdomains. The active site includes a HHxxDG sequence motif that sits at the bottom of a covered “canyon” formed by the two subdomains. With our first crystal structure solved in the lab, we described important movements that occur between the subdomains that may facilitate C domain catalysis.
NRPS tailoring domains
Tailoring domains in NRPSs allow products to access far more chemical space and are commonly found in these synthetases. For example, cyclosporin synthetase contains methyltransferase domains; daptomycin (Cubicin) synthetase, epimerization domains; bactitracin (BACiiM) synthetase, a heterocyclization domain.
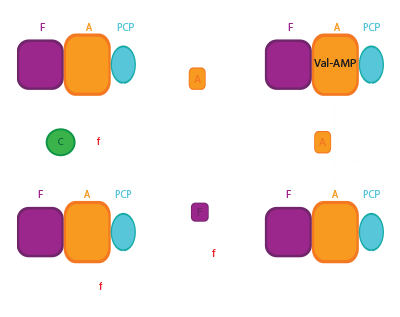

NRPS termination domains
NRPSs need specialized termination domains to release the final product. This is often the thioesterase (TE) domain. The TE catalytic mechanism proceeds through two half reactions: In the first step, the linear substrate made by the enzyme is transferred from the final PCP domain to the TE domain active site serine, forming a covalently attached acyl-enzyme intermediate.
Large NRPS proteins
In their assembly-line logic and complicated catalytic cycle, NRPS domains and modules must work in concert to synthesize the nonribosomal peptide product. To investigate the holistic workings of NRPSs, we use X-ray crystallography, chemical biology tools, biophysical techniques and biochemical methods to elucidate their structures and functions.

Cyanophycin synthesis and degradation
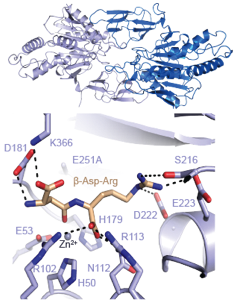

The ribosome (past research)
The ribosome is the cell’s protein factory. It translates the genetic information in mRNA into protein, rapidly and with high fidelity, using aminoacyl-tRNAs as substrates. A large number of accessory protein factors are necessary for in vivo protein synthesis, and the interplay between these factors and the ribosome is extremely complex. Deregulation of protein synthesis in humans in associated with cancers, and many important antibiotics target the bacterial ribosome.