Large NRPS proteins
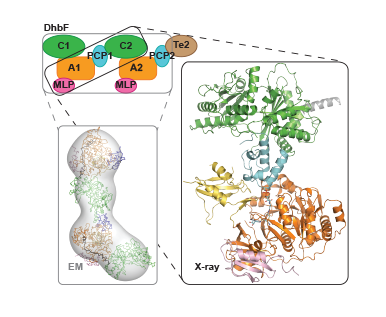
We combined electron microscopy and X-ray crystallography to get even wider views of NRPSs in action. Our recent EM studies on a dimodular bacillibactin synthetase protein produced the first 3D views of a multi-modular NRPS. Accompanying pan-module X-ray structures begin to show the higher-order architectural features NRPSs require for to perform their syntheses. Further studies of large NRPSs by EM and X-ray crystallography are a major initiative in the lab.
Relevant Schmeing lab papers: Fortinez et al, Nature Comm 2022; Reimer et al, Science 2019; Reimer et al, Curr Opin in Struct Biol. 2018; Tarry et al, Structure, 2017; Reimer et al, Nature, 2016; Reimer et al, Acta D, 2016; Tarry et al, PEDS, 2015.